
I am fascinated by research questions that lie in the sweet spot where ecology and evolution overlap in aquatic systems.
Rapid responses of marine bacteria to environmental change
The diversity of mechanisms regulating the responses of oligotrophic marine bacteria to environmental change is vastly unexplored. Microorganisms respond and adapt to dynamic environmental stimuli and growth conditions through the tight coordination of transcription, translation, and protein homeostasis. These responses shape the ecology and evolution of marine microbial communities and the biogeochemistry of our oceans. I aim to understand the regulation of transcription and translation in natural populations of marine bacteria.
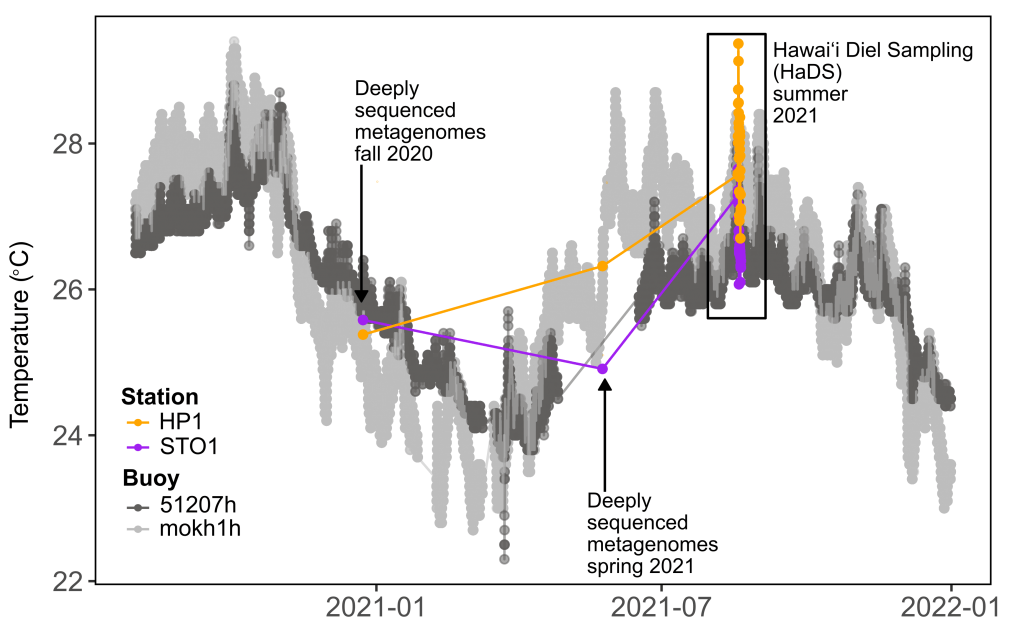
To do so, I collected parallel samples of surface ocean marine metatranscriptomes, metagenomes, and transfer RNA transcripts every 90 minutes for 48 hours through the Hawaiʻi Diel Sampling (HaDS). This represents the first sequencing of high-throughput transfer RNA transcripts from an environmental microbiome. I am utilizing novel multi-omic bioinformatic and statistical approaches to assess post-transcriptional regulation through epigenetic modifications on transfer RNAs.
Publication available here: https://doi.org/10.1038/s41597-025-06166-3
Dataset available here: https://www.bco-dmo.org/dataset/963210
Ecological differentiation of globally abundant marine bacteria
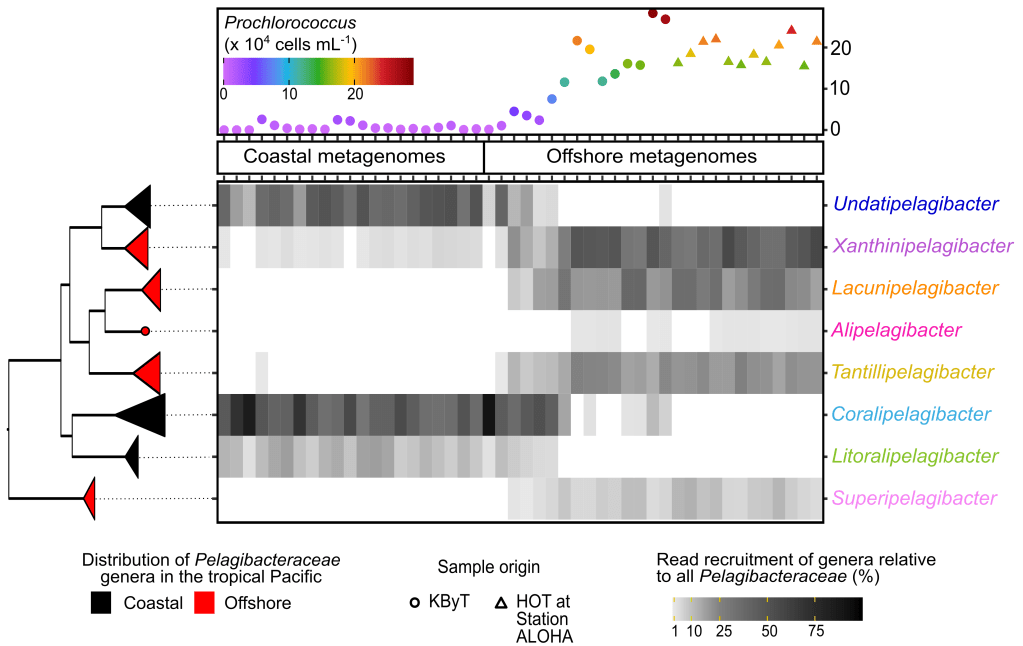
SAR11 is a free-living lineage of marine Alphaproteobacteria and one of the most abundant and ubiquitous bacterial clades on Earth. However, understanding the mechanisms and conditions responsible for ecological differentiation within this group has been difficult due to its high genetic diversity, limited number of cultivated representatives, and lack of a robust taxonomic framework.
To address this, I collaborated on three high-throughput cultivation experiments targeting oligotrophic marine bacteria. This resulted in the first cultivated representative of SAR86 and >80 new SAR11 isolates.
With the new SAR11 isolate genomes, I used comparative pangenomic approaches and metagenomic read recruitment to delineate genetic and functional variation and characterize ecotypes of the SAR11 across nearshore Kāneʻohe Bay, the adjacent offshore, and the central North Pacific Subtropical Gyre. I defined the genomic variation underlying distinct SAR11 ecologies and how these traits differ among and between SAR11 genome clusters with distributions specific to the coastal tropical Pacific and those with distributions specific to the surrounding offshore and ocean gyre waters. This work provides new insight into how SAR11’s impact on the dissolved organic matter pool and biogeochemical cycles in the ocean.
Microbes as bioindicators of ecosystem health
Microbial diversity and function are tightly coupled to environmental conditions, making microbial genomes powerful tools to monitor anthropogenic and climatic impacts on aquatic environments.
As a NOAA Margaret A. Davidson Fellow, I worked with the the Indigenous resource mangaers at the Heʻeia Fishpond and the Heʻeia National Estuarine Research Reserve, to develop a baseline understanding of phytoplankton diversity and biomass across multiple habitats that link the coastal environment of Oʻahu, Hawaiʻi, with the offshore. The increase of nutrient supply coming from both land and sea in estuarine-coastal systems can elevate rates of phytoplankton primary productivity and support increased productivity and biomass across trophic levels in the estuarine and marine food web. However, climate-change associated factors (e.g. ocean stratification, rain intensity, storm frequency) will likely impact the delivery of nutrients to phytoplankton in both coastal and offshore environments of the tropical Pacific. Ongoing restoration of Indigenous aquaculture systems located within the Heʻeia National Estuarine Research Reserve (NERR) on Oʻahu, Hawaiʻi aims to maximize herbivorous fish productivity through the aquatic food web. We also co-developed microbial biomarkers to track climate-driven changes in community composition and identified priority areas for management and restoration.
Check out our publication: https://doi.org/10.1002/lno.70075
Dataset available here: https://www.bco-dmo.org/dataset/930163
I am involved in efforts to advance, coordinate, and innovate the application of marine microbial ‘omics to inform and enhance management of ocean resources. Currently, I am co-leading the writing of a perspective piece that results from conversations during the UN Decade endorsed Marine Microbial Observatories for the Future Workshop. I also spoke at the Towards Sustainable Global Marine Omics Observation and Coordination Workshop.

Paritioning genetic diversity of marine bacteria into ecological units via time-series ocean observations
Surveys of microbial genetic diversity and environmental parameters across environmental gradients can provide a useful approach to examining the distribution of microbial populations across ecological niches and investigating the interplay of ecology and evolution.
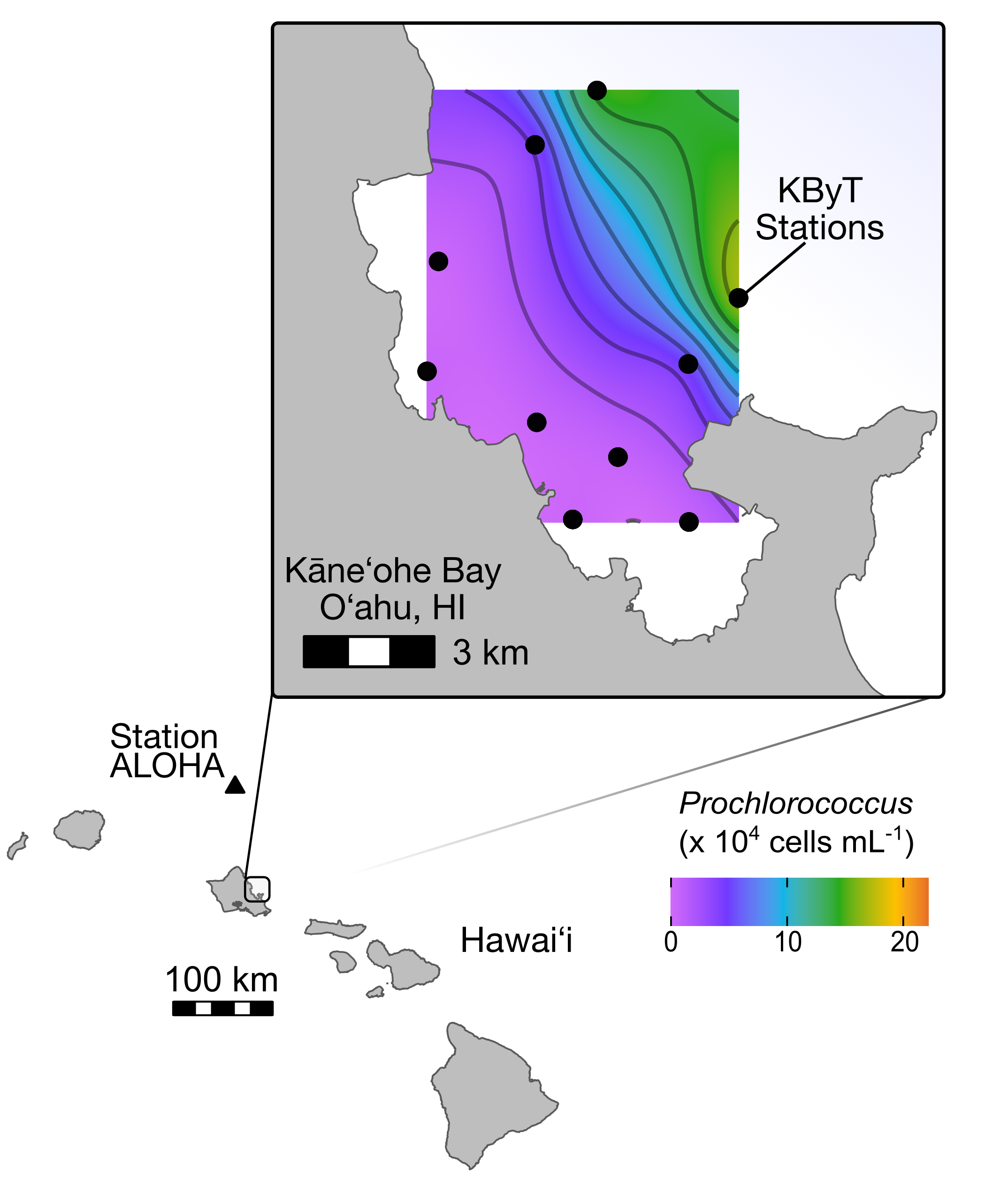
As a PhD student in Dr. Michael Rappé’s Laboratory, I helped establish the Kāneʻohe Bay Time-series (KByT) which samples surface ocean biogeochemistry and microbial communities across a coastal to offshore gradient.
I examined the spatio-temporal distributions of the 5 major clades of SAR11 marine bacteria across seasons and along the nearshore to offshore physiochemical gradients within and adjacent to Kāneʻohe Bay. This study characterizes how SAR11 genetic diversity partitions into distinct ecological units in coastal and offshore environments of the tropical Pacific, and correlates this diversity with abiotic and biotic parameters, including patterns of Prochlorococcus and Synechococcus abundance.
Publication available here: https://doi.org/10.7717/peerj.12274
Dataset available here: https://www.bco-dmo.org/dataset/930084
Diversity and activity in marine microbial eukaroytes
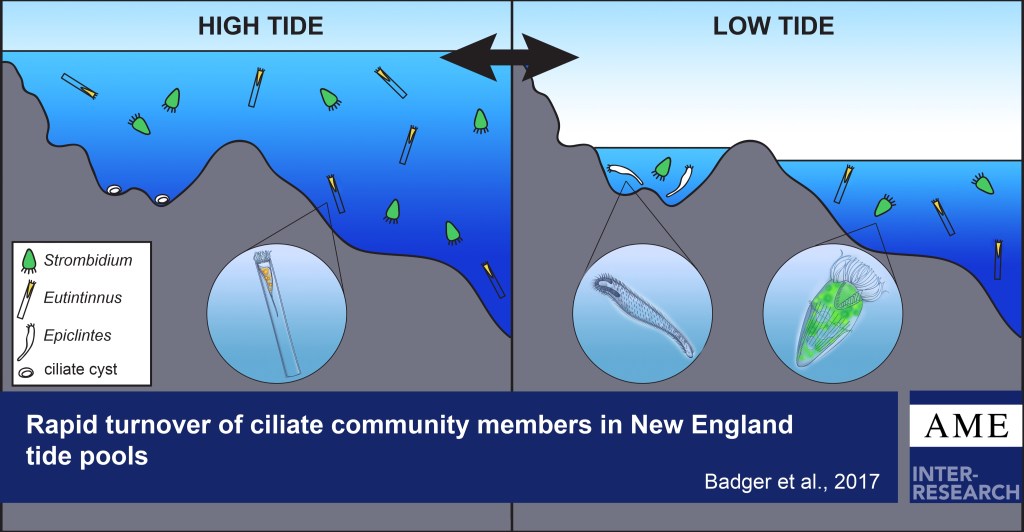
Planktonic marine microbes play important roles in both biogeochemical cycling pathways and as a links between bacteria and higher trophic levels. Despite their important role of linking microbial and classic marine food webs, data on biogeographical patterns of microbial eukaryotic grazers are limited.
I examined the diversity and activity of marine ciliates in New England tide pools under different tidal conditions. I also examined the distribution of planktonic ciliates in coastal and slope waters off New England.
Marine biodiversity and capacity building in the Indo-Pacific
For nearly two years, I worked as a researcher at the Indo-Pacific Biodiversity Research Center in Indonesia, a laboratory established to advance educational and scientific capacity in Indonesia. I became fluent in Bahasa Indonesia and used these skills to train scientists from multiple Indonesian institutions in laboratory techniques, co-facilitate workshops in genetic analysis, provide feedback on grant proposals, and share insight into educational and research opportunities abroad. I co-mentored three undergraduate research theses while conducting research on grouper fish diversity and phylogeography in the Indo-Pacific.
Indonesia is one of the top grouper fish producers in the world and grouper are a main component of the growing Asian live-reef fish and luxury seafood trade. In this research, I used genetic and morphological approaches to describe two new species of grouper, Epinephelus kupangensis and Epinephelus craigi.

